Epigenetics plays an important role in the development of many metabolic diseases and since these alternations to the underlying DNA are reversible, restoring the DNA to normal provides opportunities for developing pharmacotherapies for many metabolic diseases.
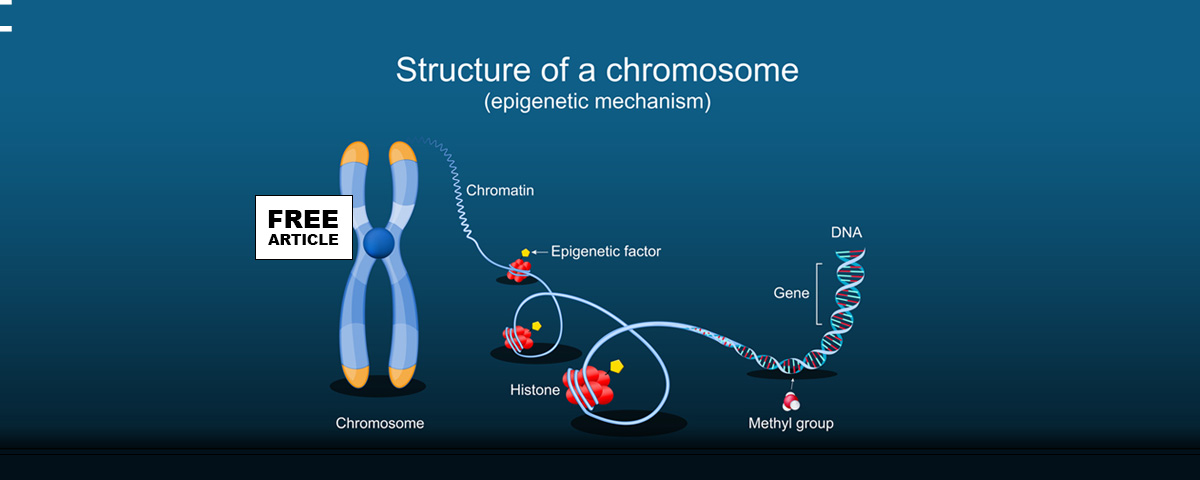
Epigenetic changes are known to be inherited changes in gene expression that take place without changing the DNA sequence underneath. In simpler terms, it is about how gene expression can be turned on or off or dialed up or down without altering the genetic code itself. These changes are stable, heritable, and carry over to cell division, allowing the spread of cell lineages to be altered. Epigenetic changes can be caused by at least three mechanisms
- DNA Methylation
- A methyl group (–CH₃) is added to DNA, typically at cytosine bases.
- This usually silences gene expression. Gene Expression is the process by which the information encoded in a gene is used to direct the assembly of a functional product, typically a protein.
- Example: In some cancers, tumor suppressor genes are methylated and turned off
- Histone Modification
- DNA wraps around proteins called histones.
- These histones can be chemically modified e.g., methylation(–CH₃) acetylation (−COCH 3).
- These modifications affect how tightly DNA is wound and, in turn, whether genes are accessible for transcription. Transcription is the process by which the information encoded in a segment of DNA is copied into messenger RNA (mRNA). This is the first step of gene expression, leading to the production of proteins.
- Non-coding RNAs
- Small RNAs micro-RNA (miRNAs) can interfere with messenger RNA (mRNA) and affect whether genes are translated into proteins.
- Some non-coding RNAs help guide other epigenetic modifications.
In summary, increased DNA methylation at gene promoters causes decreased gene expression, increased non-coding ncRNA activity reduces gene expression through degradation of messenger RNA (mRNA), while the effect of histone modifications on gene expression is more variable depending on the type and location of substrate attachment to the histone tail.
ENDOMETRIOSIS
Endometriosis is a chronic condition where tissue similar to the uterine lining grows outside the uterus. While genetics play a role epigenetic modifications are crucial in altering gene expression without changing the underlying DNA sequence. These epigenetic changes help explain key features of endometriosis such as estrogen dominance, progesterone resistance, chronic inflammation and invasive endometriotic lesion behavior.
Several studies reported a significant difference between specific genes’ methylation levels in endometrial biopsies and normal tissue, which suggests that DNA methylation may play an important role in the modulation of the genotype in endometriotic tissue. These studies identified an association between endometriosis and hypermethylated genes, including the PGR-B, SF-1, and RASSF1A. The genes HOXA10, COX-2, IL-12B, and GATA6 were found to be hypomethylated in endometriotic tissue by several studies. Acetylation and methylation are the two key histone modifications leading to differential gene expression in endometriotic tissues.
PCOS
Polycystic Ovary Syndrome (PCOS) is a complex endocrine disorder affecting 6–20% of reproductive-age women worldwide. While genetics contributes to PCOS, it doesn’t fully explain its variability. Epigenetic changes—heritable but reversible modifications that affect gene expression without altering DNA sequence—are now recognized as critical in its development and progression.
EPIGENETIC ALTERATIONS IN KEY PCOS PATHWAYS
| Pathway | Affected Genes | Epigenetic Change | Result |
| Androgen synthesis | CYP17A1, CYP19A1 | Hypomethylation | Ý Testosterone, estradiol |
| Insulin resistance | INSR, IRS1, PPARγ | Mixed hypo-/hypermethylation | Altered insulin signaling |
| Folliculogenesis | FSHR, AMH | Hypermethylation | Ovarian dysfunction |
| Inflammation | TNF-α, IL-6 | Hypomethylation | Chronic low-grade inflammation |
| miRNA regulation | miR-93, miR-21, miR-145 | Dysregulated expression | Metabolic and reproductive imbalance |
DNA methylation is the most widely studied epigenetic modification in PCOS. At present, changes of DNA methylation have been found in serum, ovarian, hypothalamus, skeletal muscle, adipose tissue of PCOS patients, and these changes are closely related to insulin resistance, lipid metabolism and follicular development of PCOS.
Developmental Origins (Fetal Programming Hypothesis)
Recent studies have demonstrated that pre-natal exposure to excess testosterone (androgen) predisposes young girls to PCOS. Polycystic ovary syndrome is known to be caused primarily by high prenatal and postnatal testosterone, and it demonstrates a set of phenotypes opposite to those found in endometriosis. Prenatal androgen exposure may induce epigenetic reprogramming in fetal tissues, increasing PCOS risk later in life. Evidence from animal models: Prenatally androgenized (PNA) mice develop PCOS-like features. These models show altered DNA methylation in ovarian and hypothalamic genes. Human studies support the idea that in utero environment (e.g., maternal hyperandrogenism, obesity, gestational diabetes) can epigenetically modify fetal gene expression.